CONSERVE
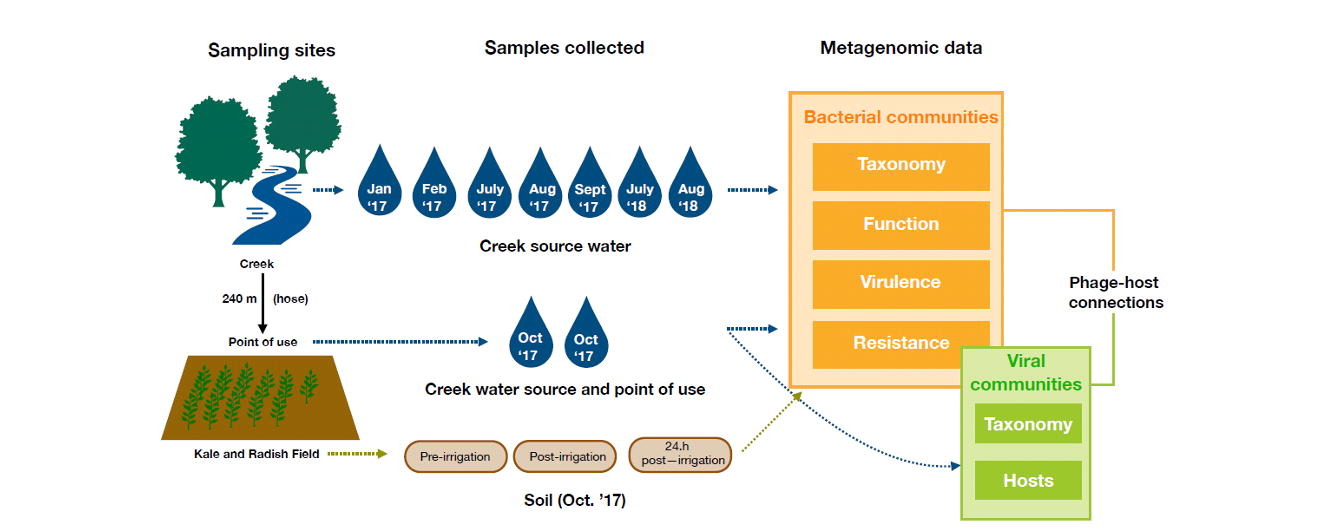
Lotic surface water sites (e.g. creeks) are important resources for localized agricultural irrigation. However, there is concern that microbial contaminants within untreated surface water may be transferred onto irrigated soil and crops.
To evaluate this issue, water samples were collected between January 2017 and August 2018 from a freshwater creek used to irrigate kale and radish plants on a small farm in the Mid-Atlantic, United States. In addition, on one sampling date, a field survey was conducted in which additional water and soil samples were collected to assess the viral and bacterial communities pre- and post-irrigation.
All samples were processed for DNA extracts and shotgun sequenced and the resulting metagenomic libraries were assembled de novo and taxonomic and functional features were assigned at the contig and peptide level.
I coordinated the computational analysis of the shotgun microbial and viral metagenomes.
Role: Metagenomics and data analysis
PI: Amy Sapkota